LIFETimes
Light-Induced Function: from Excitation to Signal through Time and Space
ERC-2017-ADG (n. 786714)
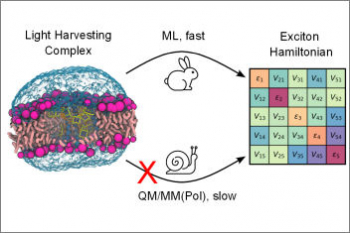
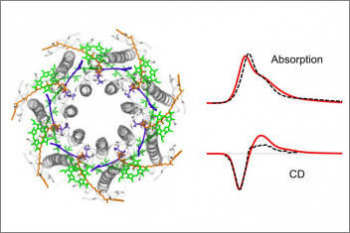
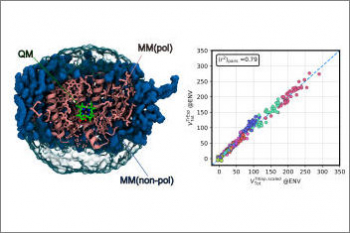
 Organisms of all domains of life are capable of sensing, using and responding to light. The molecular mechanisms used are diverse, but most commonly the starting event is an electronic excitation localized on a chromophoric unit bound to a protein matrix. The initial excitation rapidly “travels” across space to be converted in other forms of energy and finally used to complete the biological function. The whole machinery spans many different space and time scales: from the ultrafast electronic process localized at the subnanoscale of the chromophoric units, through conformational processes which involve large parts of the protein and are completed within micro- to milli-seconds, up to the activation of new protein-protein interactions requiring seconds or more. Theoretically addressing this cascade of processes calls for new models and computational strategies able to reproduce the dynamics across multiple space and time scales. Such a goal is formidably challenging as the interactions and the dynamics involved at each scale follow completely different laws, from those of the quantum world to those of classical particles. Only a strategy based upon the integration of quantum chemistry, classical atomistic and coarse-grained models up to continuum approximations, can achieve the required completeness of description. This project aims at developing such integration and transforming it into high-performance computing codes. The completeness and accuracy reached by the simulations will represent a breakthrough in our understanding of the mechanisms, which govern the light-driven bioactivity. Through this novel point of observation of the action from the “inside”, it will be possible not only to reveal the ‘design principles’ used by Nature but also to give a “practical” instrument to test “in silico” new techniques for the control of cellular processes by manipulating protein functions through light.
Organisms of all domains of life are capable of sensing, using and responding to light. The molecular mechanisms used are diverse, but most commonly the starting event is an electronic excitation localized on a chromophoric unit bound to a protein matrix. The initial excitation rapidly “travels” across space to be converted in other forms of energy and finally used to complete the biological function. The whole machinery spans many different space and time scales: from the ultrafast electronic process localized at the subnanoscale of the chromophoric units, through conformational processes which involve large parts of the protein and are completed within micro- to milli-seconds, up to the activation of new protein-protein interactions requiring seconds or more. Theoretically addressing this cascade of processes calls for new models and computational strategies able to reproduce the dynamics across multiple space and time scales. Such a goal is formidably challenging as the interactions and the dynamics involved at each scale follow completely different laws, from those of the quantum world to those of classical particles. Only a strategy based upon the integration of quantum chemistry, classical atomistic and coarse-grained models up to continuum approximations, can achieve the required completeness of description. This project aims at developing such integration and transforming it into high-performance computing codes. The completeness and accuracy reached by the simulations will represent a breakthrough in our understanding of the mechanisms, which govern the light-driven bioactivity. Through this novel point of observation of the action from the “inside”, it will be possible not only to reveal the ‘design principles’ used by Nature but also to give a “practical” instrument to test “in silico” new techniques for the control of cellular processes by manipulating protein functions through light.
LIFETimeS page on OpenAIRE
https://explore.openaire.eu/search/project?projectId=corda__h2020::034d86772f0aa47ce92c3c8aeb33c481